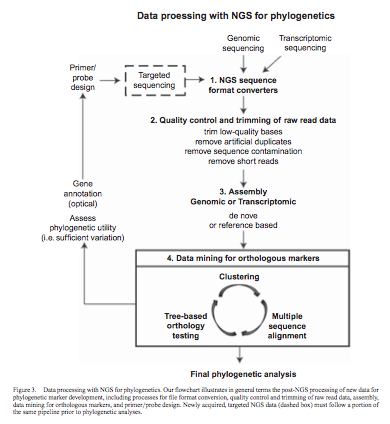
Our paper outlining how next-generation sequencing can be used in phylogenetics research has just beenpublished in Plant Ecology and Diversity. Grant Godden, Ingrid Jordn-Thaden et al review approaches and considerations that take into consideration the limited budgets of most evolutionary biology labs.
Grant T. Godden , Ingrid E. Jordon-Thaden , Srikar Chamala , Andrew A. Crowl , Nicolás García , Charlotte C. Germain-Aubrey , J. Michael Heaney , Maribeth Latvis , Xinshuai Qi & Matthew A. Gitzendanner (2012): Making next-generation sequencing work for you: approaches and practical considerations for marker development and phylogenetics, Plant Ecology & Diversity, 5:4, 427-450
See the article on the publisher’s site. If you do not subscribe to this journal, please email me and request one of the limited number of free reprints I can distribute.
Abstract
Recent reviews are setting the stage for the use of next-generation sequencing technologies in phylogenetic applications. However, the processes for developing new markers for phylogenetic analyses remain difficult to navigate for many researchers in plant systematics. We review several experimental approaches and practical considerations for developing new phylogenetic markers with next-generation sequencing technologies. We also outline a flexible framework for data acquisition that is readily adaptable to the needs of individual researchers and carefully consider cost-related issues that may be of concern to many laboratories in evolutionary biology. The next-generation and targeted sequencing approaches presented here offer considerable savings of time and money over the traditional PCR and Sanger sequencing approaches currently used in plant systematic research, particularly in cases involving large numbers of taxa and phylogenetic markers. Even with a limited research budget, next-generation sequence data can accommodate exploration of biological questions in ways that were not previously possible.